Feasibility of MinION Nanopore Rapid Sequencing in the Detection of Common Diarrhea Pathogens in Fecal Specimen | Analytical Chemistry

A computational strategy for rapid on-site 16S metabarcoding with Oxford Nanopore sequencing | bioRxiv
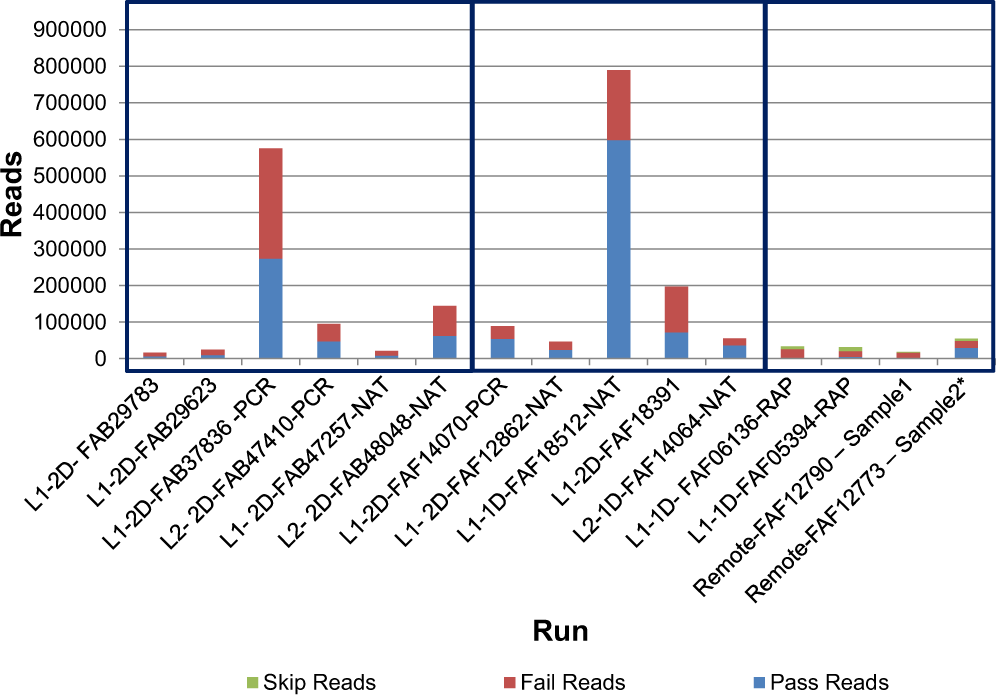
Evaluation of Oxford Nanopore's MinION Sequencing Device for Microbial Whole Genome Sequencing Applications | Scientific Reports
GitHub - nanoporetech/epi2me-api: API for communicating with the EPI2ME Platform for nanopore data analysis. Used by EPI2ME Agent & CLI.
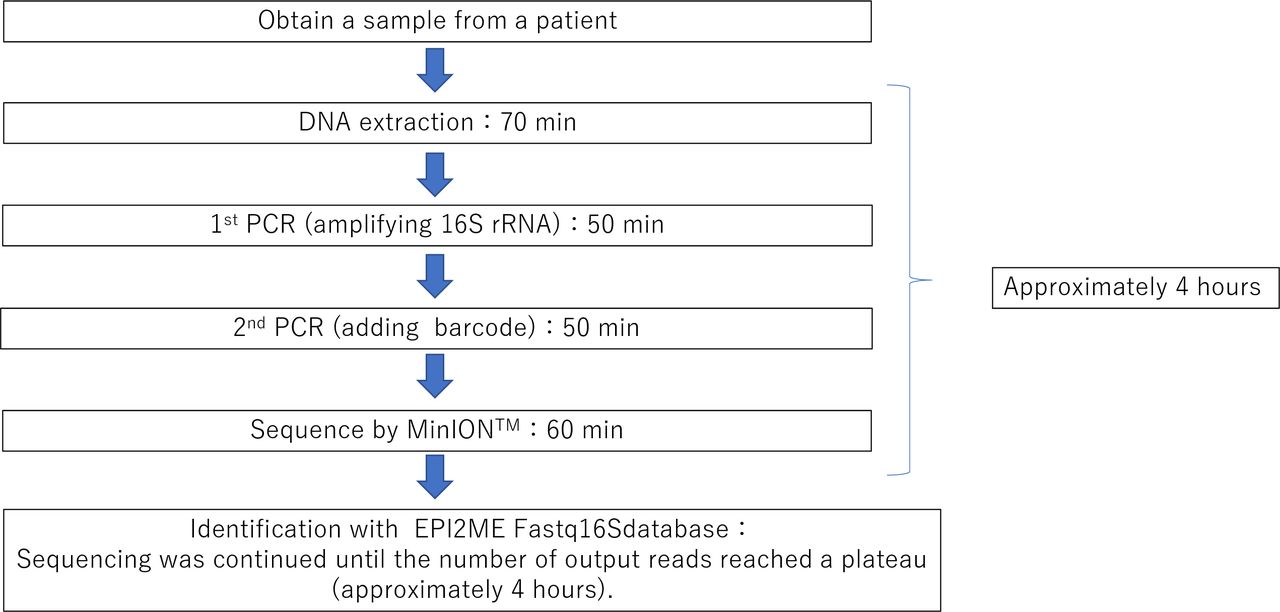
16S rRNA nanopore sequencing for the diagnosis of ocular infection: a feasibility study | BMJ Open Ophthalmology